Here at Virginia Happy Smiles Inc, we like to periodically review the latest research that is currently available to the profession of dentistry. This is a summarization of a peer-reviewed publication found here. The summarization was done by Umair Akbar. The non-profit, Virginia Happy Smiles, Inc., was founded by Hira Ansari with the goal of serving the citizens of Richmond, Virginia, for the foreseeable future.
Anatomical barrier sites are subject to frequent viral infection. Antiviral immunity at these surfaces is largely regulated by the production of interferons (IFNs) that function as critical antiviral cytokines by restricting viral infection, replication, and release. Here, we show that oral epithelial cells respond to viral agonists by preferentially inducing Type III IFNs (IFN-λ).
Porphyromonas gingivalis (Pg), an endogenous oral pathogen, dampens all aspects of interferon (IFN) signaling in a manner that is strikingly similar to IFN suppression employed by multiple viral pathogens
- In vitro, Pg suppressed IFN production by down-regulating several IFN regulatory factors (IRFs 1, 3, 7, and 9), proteolytically degrading STAT1 and suppressing the nuclear translocation of the ISGF3 complex, resulting in profound and systemic repression of multiple IFN-stimulated genes.
- Pg-induced IFN paralysis was not limited to murine models but was also observed in the oral tissues of human periodontal disease patients.
The oral mucosal barrier is heavily colonized by a complex microbiome composed of several bacterial and fungal species that interface with oral epithelial cells.
- Dysbiosis caused by specific communities of oral pathogenic bacteria that actively subvert immune pathways and suppress cytokine and chemokine generation leads to pathogenic persistence, chronic inflammation, and enhanced susceptibility to secondary infections (1, 2, 3).
- Interferons (IFNs) are critical antiviral cytokines that are essential in all aspects of antiviral immunity
- Type III interferons have emerged as the “guardians” of anatomic barriers
- The microbial colonizers at anatomic barrier sites can either stimulate or suppress IFN responses and ISG expression and thereby influence host susceptibility to viral infection
- Detection of bacterial ligands by epithelial Toll-like receptors (TLRs) can stimulate IFN-λ expression in a manner that reinforces epithelial barrier integrity
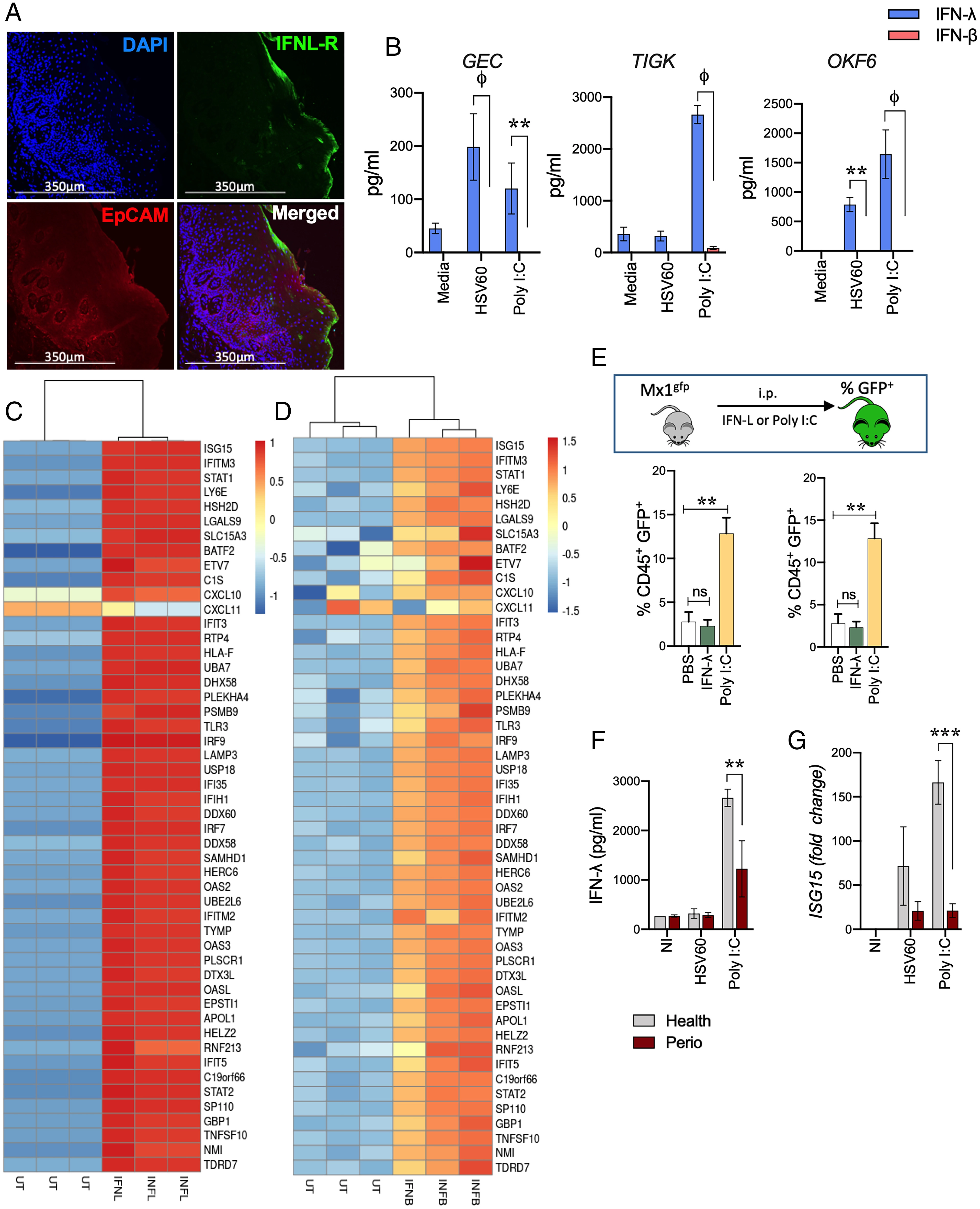
Oral Mucosal Epithelial Cells Preferentially Induce IFN-λ in Response to Viral Agonists
- GECs were isolated from human gingival tissues and challenged in vitro with poly I:C, a synthetic double-stranded RNA analog and TLR3 agonist, as well as a 60 base pair oligonucleotide sequence derived from HSV-1 (HSV60) recognized by cytosolic DNA sensors.
- Similar to the respiratory and intestinal epithelium, GEC expressed EpCAM and strongly expressed epidermal cell adhesion molecule (EpCAM)-positive region, which is located near the apical margin of the epithelial cell apical wall, and responsive to viral pathogen-associated molecular patterns (PAMPs).
- Significantly higher levels were also observed with human oral keratinocytes (OKF-6 cells) that line buccal surfaces of the mouth and also in the telomerase immortalized Gingival keratinocyte (TIGKs) cell line (30) (Fig. 1B).
Periodontal Disease and Associated Dysbiosis
- A significant predisposing factor for viral infections, or reactivation of latent viruses, in the oral cavity is chronic inflammation associated with periodontal disease (32-34).
- Pg infection inhibits IFN-λ induced by multiple viral PAMPs and PRRs such as poly I:C (TLR3 agonist), the TLR-7 agonist ORN06 (single-stranded RNA analog), and also 2’3′-cGAMP (stimulator of interferon genes [STING] agonist]
- The cyclic GMP-AMP synthase (cGAS) and STING pathway is central to recognition of cytosolic DNA from DNA viruses such as HSV-1 (38, 39)
- We also tested other Pg strains such as W83, another common laboratory strain of Pg, as well as the clinical isolate MP-504, and found these strains also strongly inhibited IFN responses
IRF-1 Regulates Cell-Intrinsic Antiviral State by Maintaining Basal Expression of ISGs
- IRFs are a family of transcription factors that play critical roles in several aspects of host antiviral immunity.
- During an infection, viral agonists induce IFN production, which via paracrine and autocrine signaling, reinforces antiviral defenses by driving ISG expression to levels several-fold higher than those observed in the basal state.
P. gingivalis Transcriptionally Represses IFNL1 by Up-Regulating ZEB1
- Multiple transcription factors, including IRF family members (IRF-3 and IRF-7), Nuclear Factor kappa-light-chain-enhancer of activated B cells (NF-κB), and Activator Protein 1 (AP-1), can all cooperatively induce Type I and Type III IFN transcription independent of IRF (42, 43, 48).
- Pg infection down-regulated IRFs 3, 7, and 9 that bind to PRD1 and ISRE elements on IFNF1 promoter, respectively.
- We focused on NF-κBs because they could potentially contribute to IFN-λ production in response to HSV60
- SerB, a serine phosphatase produced by Pg, dephosphorylates the serine 536 residue on the RelA subunit of NF -κB, preventing its nuclear translocation and blocking subsequent expression of such as IL8/CXCL8 (51).
P. gingivalis-Mediated Cleavage of IFN Receptors Enforces IFN Paralysis
- Pg produces several cysteine proteases, the arginine-specific gingipains A and B (RgpA and RgpB), that proteolytically degrade cellular proteins and attenuate signal transduction pathways.
- Although Type I and III interferons bind to distinct receptors, they activate overlapping downstream signaling pathways leading to ISG expression (14).
- GECs incubated with recombinant IFN-λ rapidly phosphorylated STAT1, however, Pg infection blocked STAT1 phosphorylation.
Discussion
- Pg infection suppressed the activation of multiple IFN-driving transcription factors such as IRFs 1, 3, 7, and 9, NF-κB, and STAT-1, subsequently blocking ISGF3-driven ISG expression.
- The concerted action of multiple Pg virulence factors blocked IFN pathways using molecular strategies that are similar to those used by certain viruses
- In our model, IFN paralysis was further reinforced by Pg via proteolytic degradation of INFL-R and IFNAR making GECs insensitive to exogenous IFNs produced by other cells such as plasmacytoid dendritic cells
Materials and Methods
- P. gingivalis, F. nucleatum, and S. gordonii were cultured in trypticase soy broth (TSB) supplemented with hemin (5 μg/mL) and menadione, and 1 mg/mL yeast extract.
- All strains were grown anaerobically (85% N2, 10% H2, and 5% CO2) at 37 °C.
Processing of Human Gingival Tissues
- Deidentified gingival tissue specimens were obtained from healthy individuals, either undergoing periodontal surgery or obtained from the internet
Immunofluorescence and Confocal Laser Scanning Microscopy
- GECs were grown on glass coverslips in 24-well plates.
- Cells were fixed in 4% paraformaldehyde for 10 min and permeabilized with 0.2% Triton X-100 for ten min at room temperature (RT).
- After blocking with 5% bovine serum albumin (20 min), cells were incubated with either anti-human IRF-1 (Cell Signaling Technologies) primary antibody 1:200 dilution (0.165 μg/mL) overnight at 4 °C and Alexa Fluor 488-conjugated anti-rabbit secondary antibody at 1:1,000 dilution for 1 h
- Slides were washed three times with PBS and then incubated further with Alexa FLAVOR-488 antibody for 1h
- slides were counterstained with DAPI (4’6-diamidino-2-phenylindole) and mounted using ProLong antifade mounting media
- Images were visualized using LAS X Life Science software (Leica Microsystems) and analyzed using Imaris software (OXFORD instruments)
- Transcript expression was determined in formalin-fixed paraffin-embedded gingiva tissues using transcript-specific probes and RNAscope Fluorescent Multiplex Assay version 2 kit (Advanced Cell Diagnostics Inc.) as per the manufacturer’s
Viral Assays
- To determine if Pg infection influenced viral replication, GECs were seeded in 24-well plates at 80% confluency.
- Cells were infected with Pg WT or the gingipain-deficient mutant at MOI of 10 for 5 h
- After a 1 h adsorption period, the inoculum was removed, and the cells were washed twice with 1× PBS to remove unbound viral particles and incubated at 37 °C in a humidified incubator in the presence of 5% CO2
- For assay, cells were examined at 24 h postinfection via a GFP-capable epifluorescence microscope
Further reading in this issue
- Emotions coordinate our behavior and physiological states during survival-salient events and pleasurable interactions
- Even though we are often consciously aware of our current emotional state, such as anger or happiness, the mechanisms giving…
- Neurobiological evidence informing about strategies to reduce xenophobic sentiment and foster…
- In sophisticated auditory-motor learning such as musical instrument learning, little is understood about how brain plasticity develops over time and how the related individual variability is reflected in the neural architecture.
- The auditory and motor neural systems are closely intertwined, enabling people to carry out tasks such as playing a musical instrument whose mapping between action and sound is extremely sophisticated.
References IFN-λ is strongly expressed in gingival tissues, cell lines, and activates ISG expression in oral tissues
- IFNL-R expression was determined by immunofluorescent staining.
- Oral epithelial cells and TIGKs were stimulated with 5 μg/mL HSV60 or 50 mg/mL poly I:C for 24 h and measured by ELISA, with data shown as mean ± SD and statistical differences determined by two-way ANOVA with Holm-Sidak multiple comparison test (**P < 0.01; ϕP > 0.001).
- Infection with Gingivalis causes IFN paralysis, characterized by the loss of basal and inducible IFN responses and corresponding down-regulated genes.
At Virginia Happy Smiles, we appreciate routinely reviewing the most recent research accessible to the dental industry. This is a summary of a peer-reviewed publication found here. Umair Akbar performed the summary above. Hira Ansari established Virginia Happy Smiles, Inc. with the intention of helping the residents of Richmond, Virginia for the foreseeable future.
Media Contact
Company Name: Virginia Happy Smiles
Contact Person: Hira Ansari
Email: Send Email
Phone: 8325108882
City: Richmond
State: Virginia
Country: United States
Website: https://www.vahappysmiles.org/
